How to Use the Autosomal DNA Segment Analyzer
About the Autosomal DNA Segment Analyzer (ADSA)
The Autosomal DNA Segment Analyzer (ADSA) is a tool that takes your data from Family Tree DNA or GEDMATCH and constructs tables that include match and segment information as well as a visual graph of overlapping segments, juxtaposed with a customized, color-coded In-Common-With (ICW) matrix that will permit you to triangulate matching segments without having to look in multiple spreadsheets or on different web pages. Additional information, such as ancestral surnames, suggested relationship ranges, and matching segments and ICWs on other chromosomes is provided by hovering over fields on the screen. You may also generate emails to persons you match from the page. The web page produced by this program does not depend on any other files and may be saved as a stand-alone .html or .htm file that will function locally or offline in your browser. You can also email the saved report as an attachment. If you want to quickly see what this does, you can try out a working sample output for a single chromosome, but the default option is for all chromosomes to be displayed.
I imagine there are quite a few people out there who don't know how to use a spreadsheet, and maybe this tool can be helpful to them as an alternative to learning Excel. I have more improvements for ADSA in mind, such as allowing you to add your own editable fields, to assign segments to named groups, record the name of the common ancestor for a group, mark segments as paternal or maternal, record notes and correspondence, etc. I am also thinking about how to support GED files.
I hope this instruction manual is helpful to you. I have tried to write it in such a way that it is useful to both beginners and more advanced genetic genealogists. If you have any comments or suggestions for improvement to this manual I would very much like to hear them. And, if you have any suggestions for improvements to ADSA or, for goodness sake, if you find any bugs please email me. Many people have used ADSA successfully, but you may run into a problem I haven't seen yet. If you find something like that, please let me know right away so I can improve the program.
Don Worth
Oxnard, California
worth@ucla.edu
Known issues:
- Support for 23andMe is not implemented yet, but planned for the future.
- The menus at the top of the ADSA pages do not function correctly. We plan to replace them at some point.
What's New in ADSA
GEDMATCH support - April 2015
A new data loader was introduced by DNAgedcom in April, 2015 that uploads data extracted from the popular 3rd party website, www.gedmatch.com. GEDMATCH is a place where people can upload their raw DNA results so that they can be compared with others who have also done so. This allows comparison of kits that were produced by different testing companies. And, currently GEDMATCH is the only way to obtain iformation about matching segments for AncestryDNA kits, since AncestryDNA does not provide a Chromosome Browser. ADSA has been modified to produce reports based on GEDMATCH data. There are a few differences between how ADSA operates with Family Tree DNA kits and how it presents GEDMATCH kits. In summary, these are:
- Certain fields that are available for Family Tree DNA kits are not presently available for GEDMATCH. These include:
Match Date
Predicted Relationship
Known Relationship
Relationship Range
Haplogroups
Surnames
Total Shared cM
Longest Block cM
So, this means that using these for sorting, selection, highlighting or display purposes may not have the results you wanted because these fields are empty in a GEDMATCH kit.
- To manage processing load on GEDMATCH's servers, only the In-Common-With indicators for your top 400 matches are provided by GEDMATCH, so you will only have ICW bricks in the ADSA report for your longer segments. You can manually determine ICWs for other matches by doing a one-to-many report for one of your matches and comparing their list of matches to yours.
- Generally, there are a lot more segments in a GEDMATCH ADSA report than for Family Tree DNA. This tends to slow down the responsiveness of your browser when viewing the ADSA report. You may wish to increase the minimum segment size in ADSA to 10 cM. Not only will doing this reduce the number of non-IBD (Identical by Descent) segments that are displayed, but it will also reduce the number of matches for which thare are no In-Common-With indicators.
- To improve performance GEDMATCH excludes very close relatives (eg. siblings, parents, children) in its Matching Segment Search report, so you will not see them as matches on your ADSA report for GEDMATCH kits.
- X chromosome matches are not presently included in GEDMATCH kits.
ADSA Version 2 - July 2014
Since the original release of ADSA in January 2014 the program has been completely rewritten. Here are the new capabilities that have been added since that time:
- ADSA now accesses its data from a server-based relational database. It no longer uses CSV (comma separated value) files as its input. Using files slowed things down and made it impossible for ADSA to handle large datasets (such as Ashkenazi) because it was necessary to read every row in the file first, even if most of the rows would not be displayed in the report. Sometimes files were so large that they could not be entirely read by ADSA before its processing time limit was exceeded. All data is now accessed from the relational database on the DNAgedcom server which is loaded during the kit download process. This improves performance and also allows you to subset your data in various interesting ways.
- Kit numbers are prompted in a drop-down menu. So you no longer need to remember your kit number to use ADSA.
- There are now several different report options (including the original "Classic ADSA" report). A drop-down menu on the main screen allows you to switch between them. In addition to listing segments, you can now list matches as well. There is also an Expert Mode which allows you to build your own report by specifying the fields you want to appear on each row, the contents of pop-up windows that will appear when you hover over certain fields, the order the rows should be sorted, where table breaks should go, and, most importantly, boolean comparisons that can indicate which rows you want to select for the report or highlight. One or more of your customized report descriptions may be saved with your account for later use.
- It is now possible to change the input parameters for your report from the ADSA output page and re-run ADSA with the new inputs without returning to the main input form.
- You can include or exclude close relatives, cousins, or remote relatives in the report. This can help with visualizing ICW patterns and is one way to reduce the size of large kits so that ADSA will successfully produce a report.
- You can now highlight rows in bold-face based on your criteria. For example, you can highlight all rows whose Ancestral Surnames include one or more names, or all rows with matches after a given date. In Expert Mode you can highlight using almost any criterion you can think of.
- If a match appears on more than one row of a table, each of their ICW bricks is now colored in so that the person "matches herself" for each of her segments.
- Lists of segments and ICWs in the pop-up menus now only apply to those segments that have been selected for the report. So, if you choose to only include chromosome 13, for example, you will not see segments and ICW names listed that are associated with other chromosomes. This change was necessary to improve performance on large datasets. However, if you run the report selecting all chromosomes the output will be the same as the original ADSA and will show segments and ICWs across all chromosomes.
- The Most Distant Cousin to Display input parameter has been dropped in favor of the new close/cousin/remote relatives checkboxes. It is possible to get even more specific about which relationships to include or exclude using Expert Mode.
- For Segment reports that are sorted into tables by chromosome, ADSA no longer outputs empty table headers for chromosomes that have no segments. This is because ADSA no longer steps through all the chromosomes one by one to produce its report - it builds a query to the database based on your selection criteria and creates tables only for the data that is returned.
- The Suppress Warnings About Name Mismatches input parameter has been removed as it is no longer needed with a relational database.
- The manual has been updated to include a section with step-by-step instructions to guide you through the process of triangulation. It also provides more information about how to interpret the patterns formed by the colored ICW bricks.
If you wish you may continue run the original ADSA (Version 1) from files but it does not include any of these new features. I do plan to continue to make the original version available indefinitely.
Nomenclature For Newbies
[If you are not a beginner with Autosomal DNA tests, you can skip ahead to the next section.]
Since I will be using some "jargony" terms in the rest of this document, it might be good to define some of them first.
| Base Pairs | A location along a chromosome - one of the "ladder rungs" of the DNA helix. |
| SNP | (Pronounced "snip") Single Nucleotide Polymorphism: a particular base pair location where mutations have occurred among humans over thousands of years. 99% of base pairs are chemically identical between any two humans. The other 1% may differ and are called SNPs. |
| Segment | A consecutive sequence of DNA results (SNP "readings") that match someone else's results in the same locations on the same chromosome. A sequence of shared DNA. |
| centiMorgan (cM) | A unit of measure for the "length" of a segment of shared DNA. However, rather than simply the number of base pairs or the number of SNPs, cM incorporates the probability that a section of a chromosome will "break apart" (crossover) during recombination - the more likely the crossover, the longer the segment in cM. As such, it is a better measure to use to compare the size and significance of segments. |
| In-Common-With (ICW) | Whether any two of your matches also match each other. ICW does not imply where or how the match between your two matches occurred. (ie. it does not indicate the segment where the match occurs. Only that the two people were considered to be related by the testing company.) |
| Relational Database | A collection of data, organized in tables that are related to one another with key fields. Relational database servers may be efficiently queried by programs using selection criteria (eg. SNPS > 500 AND CM >= 7), returning only the table rows to the program that meet the criteria. |
| Kit | A collection of data associated with a person who has had an Autosomal DNA test which lists the people he matches, the DNA segments where matching DNA was found, and the In-Common-With matches between the people he matches. |
| Ashkenazi | A Jew of central or eastern European descent. More than 80 percent of Jews today are Ashkenazim. Because of the high frequency of intermarriages among Ashkenazim, they have a high degree of interrelatedness that manifests in their atDNA results with many more matches, segments and ICWs. |
| Shared cM | Total aggregate DNA shared between the person tested and a matching person across all their shared segments, expressed in cM (centimorgans).. |
| IBS | Identical By State: All matching segments are Identical By State - meaning they are identical because the SNP values match between the two persons being compared. A subset of them are also Identical By Descent (IBD). However, IBS is often used to mean non-IBD segments. IBS segments are usually due to unphased data, where the markers for the chromosome 13 you got from your mother, for example, are randomly mixed with the markers you got from your father's chromosome 13 in the DNA sequencer. Unphased data can lead to matches that jump back and forth between the two chromosomes in a pair, resulting in what looks like a long matching segment, but that is really a meaningless result. This cross-matching becomes less and less likely with longer segments.. |
| IBD | Identical By Descent: Matching segments are IBD if they represent a segment of DNA that came from an ancestor who is common to the two persons whose DNA is being compared. |
| Triangulation | The process of comparing the tested person to at least two others and verifying that all three share DNA along the same segment of the same chromosome. This may mean that all three have a common ancestor and the segment was passed down from that ancestor. |
| Triangulated Group (TG) | A group of matching segments that occupy the same portion of a chromosome, are IBD, and where the matches (persons) associated with those segments not only match the tested person, but all match one another. |
Learn more about Autosomal DNA...
- Kelly Wheaton's Beginner's Guide to Genetic Genealogy is the best overview of genetic genealogy I have ever seen!
- Video Webinars provided by Family Tree DNA.
How to Download Your Data for ADSA
ADSA works by reading and displaying match data that you download from the testing company's web site (FamilyTreeDNA.com or from GEDMATCH). FamilyTreeDNA provides a way to download some of it directly, but the only way to get all of it is to use DNAgedcom. ADSA accesses your data by finding it in DNAgedcom's relational database. So you must first copy it from either FamilyTreeDNA or GEDMATCH and cause a copy of the data to be stored in DNAgedcom's database. This is done by running either the FTDNA downloader (which actually downloads your match data from FTDNA and then loads it into the DNAgedcom database) or by running the GEDCOM loader (which allows you to paste the data you copy from GEDMATCH's reports and then loads that data into the database). Either way, you will end up with a kit (a collection of match data) in the database. When you run ADSA, you will tell it which kit in the database you want it to report from. So, you will be working with a copy of your match data that was made at a particular point in time. This has the advantage that you won't have to logon to FTDNA or GEDMATCH every time you want to generate ADSA reports, but you will need to perform the download process described above periodically to pick up new matches that may have been added to FTDNA or GEDMATCH since the last time you downloaded your kit.
If you are downloading a FamilyTreeDNA kit, read "How to Download Data from Family Tree DNA to DNAgedcom's Database". There is a Family Tree DNA Quick Start Guide for doing this as well.
If you are going to create a GEDMATCH kit, read "How to Create an ADSA (Autosomal DNA Segment Analyzer) Report from GEDmatch". Or you can read the GEDMATCH Quick Start Guide.
| Kudos where kudos are due! There would be no ADSA tool if it were not for DNAgedcom. Rob Warthen of DNAgedcom has done us all a wonderful service by gathering the match, ICW and segment data from FamilyTreeDNA. Please support DNAgedcom by using the DONATE button on its home page! Also, we are very grateful for the programming work Family Tree DNA has done to create a back-end program that allows us to gather this data quickly and efficiently. No other DNA company has been such a wonderful partner! |
If you are impatient and just want to get on with it, now that you have a kit downloaded you can select Autosomal DNA Segment Analyzer from the Autosomal Tools menu at the top of the screen and, when ADSA's input screen appears, just click the GET REPORT button. Otherwise, keep reading and we will talk about the ways you can change ADSA's inputs next.
How to Use the ADSA Input Screen
Once you have finished the download data process and your kit is in the DNAgedcom database, direct your browser to ADSA by selecting Autosomal DNA Segment Analyzer from the Autosomal Tools menu at the top of the DNAgedcom screen. Or you can go there directly with this link:
[Be sure you are on the ADSA Version 2 page and not on the older version of ADSA. The title at the top of the screen indicates which version you are using.] If you are not already logged into DNAgedcom, please login first. You should see a screen like this:
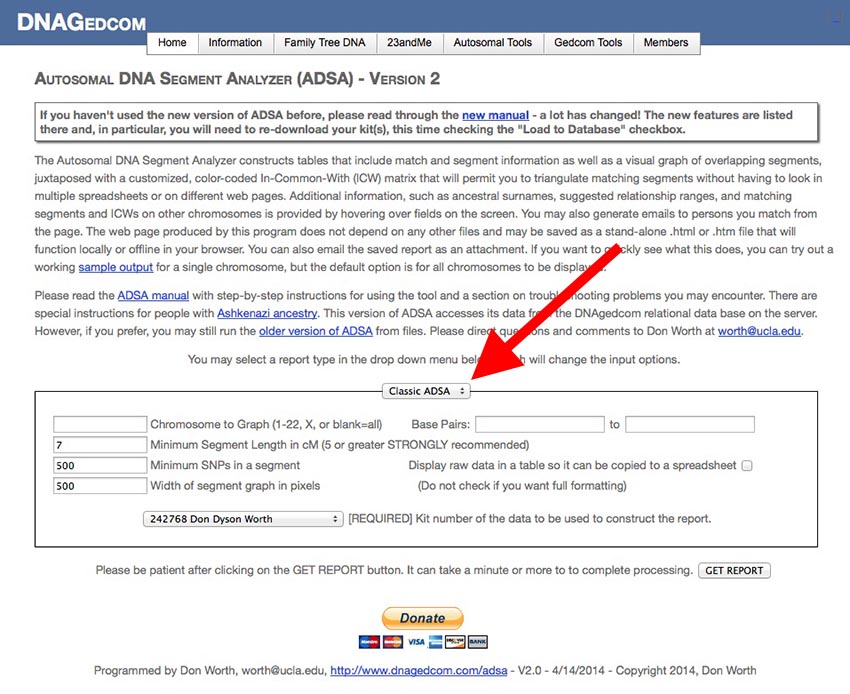
The smaller box in the center of the screen contains the input parameters you can edit to change the output report ADSA creates. At the top of the small box is a drop-down menu (my red arrow is pointing to it) with four options:
- Classic ADSA: This option runs a report of segments, sorted by chromosome number and starting and ending locations along the chromosome. This report mimics the report produced by the original version of ADSA.
- Matches: This report lists matches in one big table (segment data is not included). You can change the order in which the rows are sorted, you can include or exclude close relatives, cousins, or remote relatives, and you can highlight (in bold-face) rows with one or more surnames that appear in matches' Ancestral Surnames field or rows that have been added since a given date (or both).
- Segments: This is a variation on the Classic ADSA report but with additional input fields. In addition to the parameters from the Classic ADSA report, you can also add or exclude close relatives, cousins, or remote matches and highlight rows by surname and/or match date.
- Expert Mode: Here are the "guts" of ADSA. All of the reports above boil down to a string of text called a Report Description that defines the type of report, the fields that will appear along the row, the pop-up windows, the sort order, and the selection and highlighting criteria. You can create almost any report you can imagine in Expert Mode. There is a special section later in this document that describes how to do it.
- Database Utility: This report gives you information about all of your kits in the database. Included are row counts for each kit's list of matches, list of segments, and list of ICWs. This information can be useful in diagnosing failed download attempts (you should never see zero for the match, segment or ICW counts. If you do, you should retry the download.)
With the exception of Expert Mode, the input parameters for the other report types are described below. Other than selecting the name of the kit you want to use, most of the time you can just accept the default value for the rest of these:
Chromosome to Graph & Base Pairs
You may specify a single chromosome to graph (valid inputs are 1 through 22 or X) or leave this box blank to see segments for all 22 autosomal chromosome pairs and the X chromosome(s). The default prompt is blank - generate tables for every chromosome. If you specify a single chromosome to graph, you may also "zoom in" further by giving a starting and ending base pair position. The tool will ignore any segments that do not at least partially overlap that region on the chromosome. This is a way to reduce the size of the output file if you have so many segments that the browser bogs down looking at them (if you set minimum segment length to 1 cM, for example, or if you have Ashkenazi ancestry). If you leave the start box blank, 0 is assumed. If you leave the end box blank, 300,000,000 is assumed.
Minimum Segment Length in cM
The tool will ignore any segments that are smaller than the number you specify here. You should not go below 5 cM because the resulting page becomes so gigantic that your browser will be almost too sluggish to use. The prompt is 7 cM. (Keep in mind that some studies have found that only 30% of 7 cM segments and 5% of 5 cM segments are Identical By Descent (IBD) - the rest are false matches.) You may discover that GEDMATCH kits have so many more segments than FTDNA kits do that your browser becomes very sluggish even with a 7cM minimum, and the segment graph will be pushed off the far edge so that you must scroll to find it. It may be preferrable to set the minimum to 10 cM - especially since, for GEDMATCH kits, you won't have ICWs for the shorter segments anyway.
Minimum SNPs in a segment
The tool will ignore any segments with fewer than the number of Single-Nucleotide Polymorphisms (SNPs - aka "snips") you specify here. The prompt is 500 and that is usually the best choice.
Width of Segment Graph in Pixels
The wider you go, the easier it is to see the segments spread out across the length of the chromosome. However, if you go too wide, it may be difficult to print the page without having the browser chop things off at the right margin. The default here (500 pixels) seems to work pretty well in most cases for me (although I might have to print in landscape when I have a lot of segments on one chromosome). Once in a while you may want to make the segment graph smaller if you have a chromosome with over 50 segments on it since the ICW matrix will tend to push the table out beyond the right margin for printing.
Display raw data in a table so it can be copied to a spreadsheet
If you want to capture the segment data and ICW matrix to use in a spreadsheet, check this box. With the normal ADSA output (this box unchecked), the ICW matrix is generated with web graphics and cannot be copied to the clipboard. Also, some of the fixed width formatting of the text table to its left causes problems with copy and pasting data into a spreadsheet. Be warned, if you check this box you will just get a big table full of text and numbers and X's - no hovering pop-up windows, graphing or colors. (Note: blank/empty names and email addresses will be replaced with a question mark to preserve the columns in Excel.) The default is to leave this box unchecked so that the fully formatted, colored tables and graphs are generated.
Include Relatives: Close, Cousins, Remote
[This feature only works for FTDNA kits - GEDMATCH kits do not contain the data necessary to support this.] These checkboxes allow you to subset your matches to include or exclude close relatives, cousins or remote cousins. For example, if you want to exclude close relatives (anyone whose relationship range does not include the word "cousin") uncheck the "Close" checkbox. To exclude anyone whose relationship range includes the word "cousin" uncheck the "Cousins" checkbox. To exclude anyone with "remote" in their relationship range, uncheck the "Remote" checkbox. By default, if you check all three boxes you get every match (subject to the other constraints you have placed on the results in other fields on this form) - if you uncheck them all you will get an error that indicates you didn't select anyone for the report.
Sort By: Name, Shared cM descending, Date descending
[GEDMATCH kits do not have the Shared cM and Date fields populated, so you cannot sort on them. This feature only works properly for FTDNA kits at present.] This input chooses the order in which Match Report rows are displayed. They may be alphabetized by (first) name, or sorted with longest amounts of shared cM first, or with most recent matches first.
Highlight Matches with these Ancestral Surnames
[Surnames data is not available for GEDMATCH kits so this field only works for FTDNA kits at the moment.] You may list surnames in this box, separated by commas. Whenever one of these names appears in the Ancestral Surnames field of one of your matches, that row will be highlighted in bold-face and the name (or portion of a name) will be displayed in bold-face red in the pop-up panel for that person. Some people have locations in Ancestral Surnames and you can search for these also. The names you give will match even if they are only part of a surname (for example, "worth" will match "worthington" and "kennelworth" and "sworthy".) Blank spaces count, even if they precede or follow the name. So " worth" will not match "Kennelworth" (which might be what you want). Case is ignored. Highlights due to matching surnames are not distinguished from each other or from highlights due to Match Dates (see below). All rows that match any of the surnames or the match date are bold-faced. (Names that appear first or last in a list of ancestral surnames for a person are kind of special cases since, as they come from the testing company, they don't always start with a space or end with a space. We hope to fix this at a later date.)
Highlight Matches that are After this Match Date (mm/dd/yyyy)
[The date of the match is only available for FTDNA kits, so this feature will not work for GEDMATCH kits at the moment.] Rows in your report for matches that were added after the date you enter will be bold-faced. Highlights due to match dates are not distinguished from highlights due to Ancestral Surname matches (see above). All rows that match any of the surnames or the match date are bold-faced.
Kit Number
This drop-down menu displays all the kits (test results) you have loaded from FTDNA or GEDMATCH to DNAgedcom's relational database. Select the one you want ADSA to use to generate its report. If you have both Family Tree DNA and GEDMATCH kits in the database you can tell them apart from the kit number which appears in front of the name. Family Tree DNA kits start with a number or the letter "B" (for kits that were transferred to FTDNA from other testing companies). GEDMATCH kits start with the letters "A" for Ancestry test results that were copied to GEDMATCH, "F" for Family Tree DNA tests, and "M" for 23andMe tests.
When you're ready, click on the GET REPORT button and the tool will query the database for your data and generate the web page. It may be just a few seconds, or it may take one or two minutes if you have a lot of In-Common-With matches and/or you have a lot of segments. If you run into trouble at this point consult the If Things Don't Work For You box at the bottom of this page.
Once you've generated a chart, the input form is included at the top of the report page. You can change any of the parameters there and rerun the report. You'll also find that using the browser's BACK button will preserve the inputs you already had entered on the original entry form, allowing you to change just the ones you want to change and re-run ADSA. If you reload the page or navigate to its web address again or change the report type, everything will return to the default settings and you will have to also re-specify your kit.
Interpreting Your Results
[If you have used ADSA previously or you are very familiar with autosomal DNA segment analysis, you can skip this section. However, in addition to the walk through of what is on the output page, I have also added an explanation of and a step-by-step guide to triangulation and some interpretive analysis of the patterns formed by ICWs. The ICW pattern discussion may be of interest even to those familiar with triangulation.]
If you run either the Classic ADSA or Segments reports, this is a sample of what you'll see (I have blurred out the names to protect the innocent):
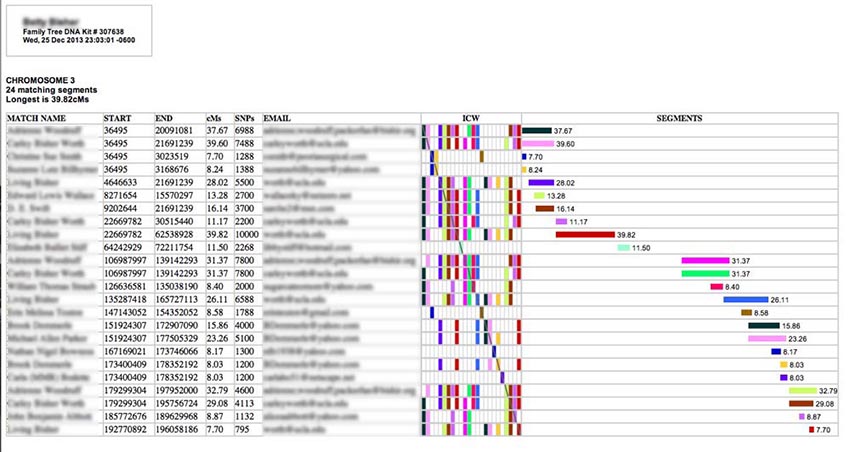
Each row describes a separate matching segment on the chromosome. The textual information in the left-hand columns describes the segment. The match person's name and email are also provided. (If a name or an email address is too long, it will be truncated - an ellipses indicates this - but you can see the full entry by hovering your cursor over it for a moment.) In the center of the table is an ICW matrix (more about that later), and on the right is a graphical representation of the matching segments spaced out along the chromosome from left to right and annotated with their length in cM (centiMorgans).
Hovering your cursor over one of the names on the left will produce a pop-up "tool tip" something like this:

In addition to the matching person's name and email address the pop-up includes the date the match occurred, the predicted relationship range, and the list of ancestral surnames. GEDMATCH kits do not have data for the match date, relatioship, or surnames, so only the name, email and GEDMATCH kit number are present in the pop-up window. If you have used the Segment report or Expert Mode to highlight surnames (FTDNA kits only), they will be displayed here in bold-face red text.
Hovering your cursor over one of the colored horizontal bars on the right side under "SEGMENTS" will produce a pop-up like this one with even more information:
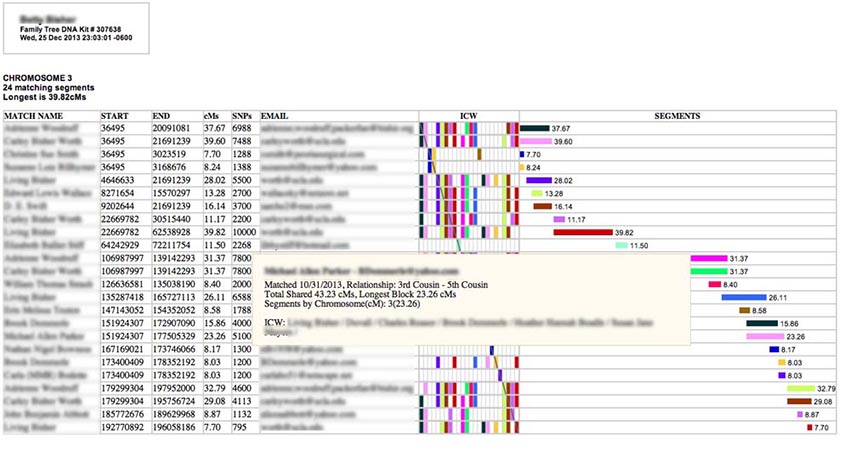
Here you will see Total Shared cMs for this person (the total length of bits of matching DNA spread across all the chromosomes), the longest block you share with them in cM (which may or may not be on this chromosome), and a list of all the matching segments you have with this person among the segments selected for the report. (In this case I had only one shared segment - and it is on this chromosome.) [GEDMATCH kits do not have total cM or longest block values, so these will be missing from GEDMATCH pop-ups.] Also listed are all the In-Common-With (ICW) names for this person among the matches selected for the report. (If you have selected all chromosomes for this report, you may see ICWs that appear with segments in other chromosome tables, while, if you have restricted your report to a single chromosome, you will only see the ICWs for people with segments on this chromosome.) A more complete explanation of ICWs is given a bit later in this manual. [GEDMATCH ICWs are only provided for the top 400 matches. So some of the rows for smaller segments may not have ICW bricks, even if two matches are ICW one another.]
If you click on one of the emails in the email column of the table, you will be transferred to your email program with the TO: line containing the match name and the email address. (Family Tree DNA does this also, but does not include a name with the email address, which, I have found, can sometimes cause an email to end up in the recipient's SPAM folder.)
The segments are plotted on the right of the ICW matrix, scaled to base pairs (so, as with the FTDNA Chromosome Browser, the apparent length of the segment may or may not be representative of the length of the segment in centiMorgans) and, to save on horizontal screen space, I did not include any empty portions of the chromosome prior to the first segment or after the last one. If there are only a few matching segments on a chromosome, this can sometimes make relatively short segments look very long, since they expand to fill the width of the table. However, the main purpose of the graph is to visualize overlaps and I was trying to avoid clipping at the right margin during printing.
The ICW matrix has been generated to include ICW information only for the persons associated with the matching segments listed for this chromosome.
WHAT IS A MATCHING SEGMENT?
Before we dig much deeper into how to interpret your ADSA report, a short digression to explain segments of matching DNA might be helpful. (If you already know the basics of segments, phased and un-phased data, IBS, and IBD, you can skip this sub-section.)
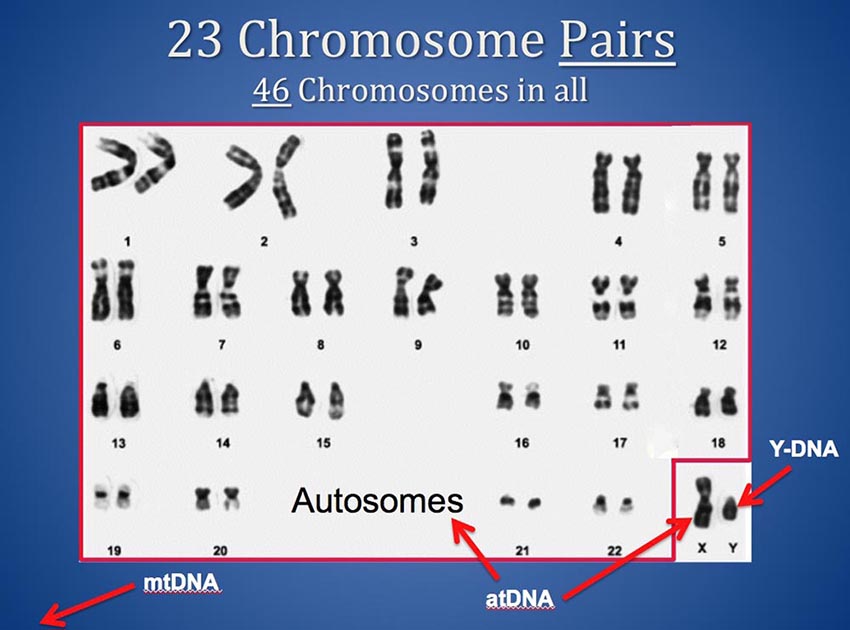
You have 46 chromosomes and they come to you in 23 pairs from your mother and father. For each numbered pair of "Autosomes" (1 through 22) you have one from your father and one from your mother. And you have a Y from your father and an X from your mother if you are male, or an X from your father and an X from your mother if you are female. (Your Mitochondrial DNA - mtDNA - comes from outside the nucleus so it is not part of this discussion.)
Thus, you actually have two chromosome 13s, and two chromosome 6s and so on. And, if you are female, you have two X chromosomes also. Most chromosome browsers plot only a single horizontal bar to depict segments that match on a chromosome - but a sequence of your DNA that matches someone else's (also known as a matching segment) can be on either chromosome of your or your match's pairs. This means two matching segments could occupy exactly the same location on a given chromosome but actually represent matches to two different common ancestors on opposite sides of your family - one in your mother's tree and one in your father's tree.
Not all matching segments are representations of a legitimate DNA match. Because most testing companies' autosomal DNA sequencing produces un-phased data (where the DNA values of your mother's and father's chromosomes at each genetic marker are not identified separately but are presented as a pair of letters in an arbitrary order at each SNP) it is possible for the equipment to find sequences of seemingly matching DNA when, in fact, matching markers are coming alternately and randomly from both chromosomes in the pair. Consider this example:
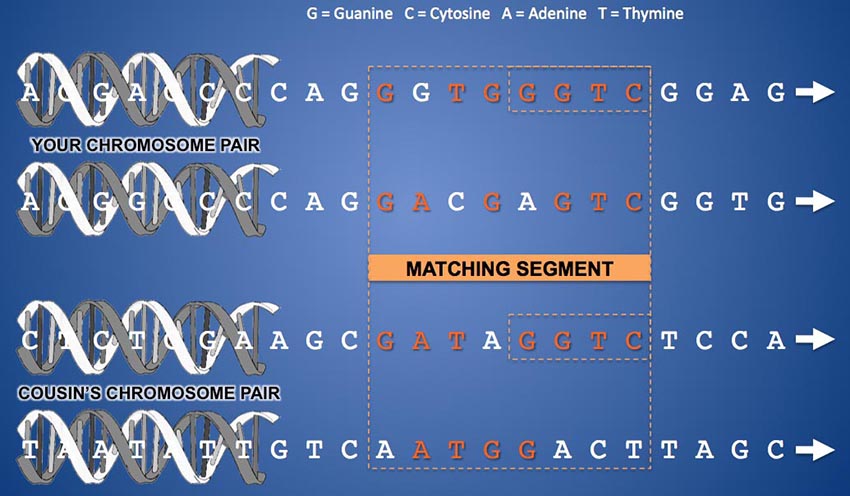
In the diagram above, let's imagine that the two chromosomes at the top are your chromosome 13 pair. And let's suppose the topmost one is from your father and the one just below it is from your mother. The "ladder rungs" that connect the strands of the double helix of one chromosome are pairs of molecules called "bases". A "rung" can be a Guanine molecule attached to a Cytosine molecule or an Adenine attached to a Thymine. Each "ladder rung" along a chromosome is called a "base pair" and is numbered from zero at the start of the chromosome to a number in the 100s of millions at the end of the chromosome, depending on the length of the particular chromosome. The vast majority of these base pairs (about 99%) are structured identically in all human beings. Only 1% of the base pair locations on a chromosome vary from one person to another. Base pairs that can differ between people are called SNPs (Single Nucleotide Polymorphisms - or "snips") and are what allows us to compare and contrast individuals' DNA. In the example above, when comparing your DNA to someone else's, the testing company compares either of the letters that appear at a given horizontal location along your two chromosomes with either of the letters that appear in the same location on the same chromosome pair of someone else's test results. For a match to be legitimate, all of the comparisons of a segment should come from the same chromosome of your pair and your predicted cousin's pair. However, you can see that, while this is the case for the last four SNPs in the example above, it is not the case for the first four. The comparison for the first half of the segment switches back and forth between chromosomes. The resulting "match" of the first four SNPs in this segment is not a match at all (in fact, this would be called a "pseudo-segment"). Only the last half of this segment is a legitimate DNA match, and even then, your two chromosomes don't have tiny "Mom" and "Dad" labels on them, so the sequencer can't tell you which side of the family the match is on.
In fact, for short matching segments, "false segments" like this (referred to as non-IBD or Identical By State (IBS), that is by haphazard chance) vastly outnumber legitimate ones (referred to as Identical By Descent (IBD), that is inherited). The longer the segment, though, the more likely the segment is due to there being an actual match along the same chromosome.
The following table (borrowed from John Walden and the International Society of Genetic Genealogy - ISOGG) illustrates this principle quantitatively:
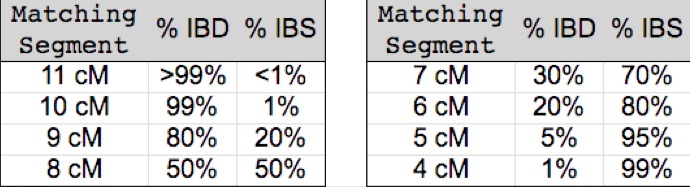
As you can see, valid (IBD) segments are few and far between among segments less than 5 cM in length. (Although some more recent research suggests that there may be more IBD segments among very small segments than previously expected - perhaps as many as 20%).
TRIANGULATION
Triangulation is a method that allows you to find groups of distant cousins who may share a common ancestor with you. Triangulated Groups (TGs) increase the likelihood that a segment is a valid match (or Identical By Descent - IBD), they separate matches along the same portion of a pair of chromosomes into those that match either one or the other of the pair, and they improve the odds of finding the common ancestor through group collaboration and the exchange of more genealogical information.
A Triangulated Group (TG) consists of two or more overlapping IBD segments that match your DNA where all three (or more) of you share the same segment of matching DNA in common. True triangulation requires knowing whether all of the members of a TG share the same segment, which would require that you have access to each person's test results. This is not possible (for privacy reasons) in Family Tree DNA. Instead, FTDNA provides In-Common-With (ICW) data which tells you whether or not two of your matches are genetically related to one another. While not quite as reliable as knowing whether they share the same segment with you and each other, most of the time, if the segment is long enough and the relationship is at 3rd cousin or more distant, ICWs serve as an adequate surrogate for true triangulation because, for more distant cousinships, there usually exists only a single segment longer than 7 cM that contributed to the genetic match.
If your autosomal DNA data was "phased" (mentioned in the preceding sub-section) then there would be no necessity to check the ICW to see if a group of people whose segments overlap have the same ancestor - they just would. The problem is that most testing companies (FamilyTreeDNA and 23andMe) produce unphased data. (AncestryDNA "pseudo-phases" its data when comparing kits within its database, however the raw data provided to AncestryDNA subscribers that can be loaded to GEDMATCH is unphased.) As we said, you actually have two chromosomes for every number from 1 to 22 - one from your mom and one from your dad. The DNA sequencer can't tell the difference between them - it just sees that there are two results for each location on chromosome 13, for example, and reports both readings in a random order. So, when your two chromosome 13s match someone else's two chromosome 13s you can't tell whether you are matching the one you got from mom or the one you got from dad. As mentioned earlier, you could, in fact, have matches to a specific area of chromosome 13 that are matching one of your two chromosomes and other matches in the exact same spot matching the chromosome you got from the other parent. So, if you look at a group of overlapping segments, some of them could be because of matches to an ancestral segment passed down from your mom's side of the family and others might be related to one of your dad's ancestors. Assuming you can't go look at someone else's test results, the only way to separate paternal and maternal matches is by using ICWs. ICWs tell you (only) that your testing company has predicted that two of your matches are related. (Although this prediction isn't always 100% accurate.) ICW doesn't say how they matched (where the match was on their chromosomes). But if you can see fairly long segments for each of them where they match you and they overlap, and the testing company says they are related (match each other), and they are not close family (a sibling or a child or grandchild to you), then they must be related to you on one side of your family (your dad's) or the other (your mom's) but not both. However, triangulation depends on the assumption that your parents are not (closely) related to one another in order to sort out the paternal and maternal chromosomes. If, however, your parents are related - you can't use triangulation quite as easily. This is particularly the issue with members of certain endogamous populations like Ashkenazi Jews where there was historically so much intermarriage in the general population that everybody is related to everyone else. More on this later.
So, what do you do about double-relationships you might have? For the purposes of triangulation, you might be OK with the fact that someone on one side of your family married someone on the same side of the family since you are trying to sort out between your mom's and dad's chromosomes. But for a situation where your mom and dad are distantly related, there could be a problem if there is any shared autosomal DNA between them. The hope is that it is a distant enough relationship that it is out of the "effective range" for autosomal DNA (beyond 5th or 6th cousin so that the odds of sharing DNA are pretty small.) My wife is my own 9th cousin once removed but we share no discernible DNA. So triangulation would work for my daughter's autosomal DNA data.
To find and exploit triangulated groups, you should follow these steps:
- Pick out a nice, long segment in your ADSA output (to be sure it is IBD)
- Look for all the segments that overlap it
- Compare all the In-Common-With (ICW) matches for the people associated with these segments to make sure everybody matches everybody else
- If all three of the above conditions are met, the "owners" of those segments probably all share a common ancestor with you and you are all part of a Triangulated Group (TG)!
- See what you can deduce about the common ancestor by looking at your matches' Ancestral Surnames
- Then contact everyone by email and ask them to help you find the common ancestor
- Repeat steps 1 to 6 for the next TG you can find
All of that said, if you prefer, you can actually perform the triangulation in a different order. If you wish, you can first look for patterns in the ICW bricks to visualize what might be TGs. Some of the ICWs form what look like rock candy crystals strung along a diagonal "string". I often start with those, then look to the right to make sure all the participants in a given "block" of ICW bricks have overlapping segments and that at least one of them is long enough to be highly likely to be IBD. As long as you meet the conditions for a TG, it's all good.
Pick Out a Nice, Long Segment
Many people choose to ignore segments under 10 cM because so many can be non-IBD. However, if you start with a long-ish segment as the first member of a Triangulated Group - something 10 cM or greater - then you can consider overlapping, somewhat shorter segments (6 or 7 cM) and still be relatively confident that they are IBD if they meet the rest of the triangulation requirements. The main problem with short segments, though, is that even if they are IBD, they generally indicate a common ancestor that is so far back in time that you and your match's paper genealogies won't go far enough back to reveal the connection. For this reason, I recommend starting with the longest segments you can find in your TGs, and spend time on the short ones when you've run out of longer ones. (Pick the low hanging fruit first!)
Another option, if you prefer to direct your genetic genealogy research to a particular line, is to search for a clearly IBD segment (10 cM or more) where a surname you are researching is listed in that match's Ancestral Surnames field. You can use the Highlight Matches With These Ancestral Surnames input parameter in the Segments report to accomplish this. [not currently supported for GEDMATCH kits.]
Look for Segments that Overlap
Overlapping means the segments occupy at least some of the same locations on the same chromosome pair. There are different schools of thought about how much of an overlap is sufficient to call two segments "overlapping". Since the overlapping portion could be IBS (non-IBD), it's probably a good thing that there be at least 7 or 8 cM in overlap (my opinion). In the ADSA output, overlapping segments are stacked up one above the other. The main reason we want to find overlapping segments, is that if two segments occupy the same space on a chromosome pair AND the owners of the segments all match each other (are predicted relatives), then you can be safe in assuming that you don't have some of the segments matching your mother's chromosome and some of them matching your father's chromosome. If everyone matches each other (and they are not your brothers or sisters or your own progeny), then they must be related to you on one side of your family or the other - hopefully not both (unless your parents are related). And, if the segments in question are long enough to represent the probable reason why the matches involved in the TG were considered matches to each other, then you can be reasonably assured that your new cousins are matching each other in the same way they match you - a common ancestor. This is why triangulation works.
Compare All of the Segments' In-Common-With (ICW) Matches
The columns and rows of the ICW matrix indicate whether or not the person named on a row matches (is predicted to be related to) another person in the table - but it doesn't (necessarily) mean that the match is because of the segment to the right (although it often is). Each person listed on the left has both a row and a column in the ICW matrix. People with names earlier in the list have columns on the left side of the matrix, while people whose names are near the bottom of the table have columns on the right side. A person's row meets his column where the diagonal line (backslash) runs from the upper left corner of the matrix to the bottom right corner.
Let's look at the example below, seven rows taken from a larger chromosome table in ADSA.

[Note: I have added the colored shading and arrows to illustrate the process - they do not appear on the ADSA output.] All of the segments on the far right of the figure above appear to overlap one another - they are "stacked" and generally occupy the same locations along the chromosome, although some are longer than others. Let's start with Bugs Bunny's segment (the yellow-green one with a length of 16.75 cM). If you follow Bugs' row to the left from his segment into the ICW matrix you will eventually arrive at the cell in the matrix where Bugs' row meets his column. This cell is marked with a backwards slash. If you then travel either up or down Bugs' column you will see that the color of the little "bricks" in his ICW column matches that of Bugs' horizontal segment on the far right (yellow-green) forming a sideways "T". (The colors are arbitrarily assigned and are meant to help connect a horizontal segment with the person's corresponding column in the ICW matrix.) Each segment that is opposite a yellow-green "brick" in Bugs' column is ICW to Bugs. That is to say, those segments belong to people who match Bugs and match you (the person tested) - they are matches you have in common with Bugs. For example, you'll notice that there are yellow-green bricks opposite the rows of Fred Flintstone, Betty Rubble, Daffey Duck, and Huckleberry Hound. All four of them are matches to you and are matches to Bugs Bunny. You, Bugs and Fred are said to be "triangulated" since there are three of you and you all match each other with overlapping segments. The same is true for you, Bugs and Betty or Daffey, or Huckleberry. Assuming the segments are long enough to be IBD (and they appear to be) and that there isn't a coincidental ICW between Bugs and Fred (and you are actually matching segments at the same location on different halves of the chromosome pair - which is rather unlikely), that means that you, Bugs and Fred all share a common ancestry. In fact, you can see that the ICW columns for Betty Rubble (dark purple), Bugs (yellow-green), Daffey Duck (brown), and Huckleberry Hound (magenta) form a multi-colored rectangle, or block of ICW "bricks". That rectangular collection of ICW "bricks" means that you all ICW each other and all of you probably share the same common ancestry. Look above Betty and you will see that Fred Flintstone also has the same configuration of colored bricks along his row as the others - he is part of the group as well. (Always look for longer overlapping segments that may be above or below the rectangular formation that could be part of a group. Their ICW rows will look similar to the ones that are part of the rectangle.) I have shaded all these ICWed rows pink. Now look at Wilma and Barney. While their segments look like they completely overlap those of the others, they do not participate in the ICW block for the pink group. In fact, they form their own little 2x2 block of bricks, meaning they match you and each other, but they do not match Fred, Betty, Bugs, Daffey or Huckleberry. Because they occupy the same place on the chromosome as the others, it's clear that they are matching the other chromosome of the pair. If the pink group matches, say, the chromosome you got from your mother, then the blue group matches the one you got from your father (or vice versa). In this case, Wilma and Barney's blue group is embedded in the larger, pink group.
When you are looking at ICW "bricks" you'll notice that almost all of the time, the patterns are symmetrical around the diagonal line. If A matches B, then B must match A. So if you see a colored brick - let's say the pink one to the left of Bugs Bunny's "slash", you can determine whose brick that is by following that column up or down until you hit a pink brick with a slash through it. In this case, that happens on Fred Flintstone's row - so Fred matches Bugs. And, on Fred's row, you will see that there is a yellow-green brick in Bugs' column. The mirror image of the pink one we started with. The symmetry is a little hard to visualize because the bricks aren't square. [Rarely, there is a brick that doesn't have a symmetrical "partner". This seems to be due to a glitch in the data coming from the testing company. My guess is that the matches for the two kits were computed at different times with slightly different match criteria.]
TIP: If you have trouble seeing the ICW cells, you can use your browser's enlarge feature (ctrl + on Windows or Command + on MacOS) to zoom in on it. I also use ctrl - (or Command - on the Mac) to shrink the screen for chromosomes with a lot of segments to see all of them on the screen at once.
Interpreting the Patterns in the ICWs
Often you will see patterns in the ICW matrix like this:

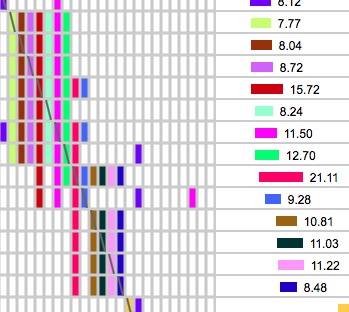
Look for small rectangles on the diagonal line of slashes like the one in the upper left that is outlined in red. That 4x4 block represents four segments belonging to people who all match each other and match you. Occasionally you will get something like the big pattern in the upper right. These big blocks of bricks are often associated with fairly short segments, so be sure the segment lengths are long enough to be IBD. (IBS segments often stack up like that - try running with minimum cM at 1 or 2 on a single chromosome and you will see a lot of them!). Remember that 50% of the time 8 cM segments are not IBD and you don't get very close to 100% certainty of IBD with segments smaller than 10 cM. However, if this big block of ICW bricks is associated with overlapping segments that are truly IBD, my guess is that the common ancestor has a LOT of descendants in the general population! I figure the more matches I can find in one Triangulated Group, the more help I will get from all of those people in finding the common ancestor. The one on the bottom is the pattern to watch out for. Clearly that brown 8.27 cM segment outlined in red that doesn't match the others is matching on the chromosome of the pair that came from my other parent. Either that or it is IBS (non-IBD).
 |
 |
 |
Sometimes you won't get a perfect ICW rectangle - there might be a few white gaps here and there - "missing teeth" as in the example above left. That can be because the corresponding segments don't overlap by enough to be considered a match to each other. Or it could be because other requirements for a match weren't quite met. For example, Family Tree DNA requires that there be a minimum of 20 cM shared DNA across all the chromosomes for two people to be declared a match, and in this case, even if the segment in your TG is long enough to be IBD on its own, your two matches may not ICW because they don't share enough total cM with each other to be considered a match. Despite the "missing teeth" in the ICW matrix, it may still be a decent Triangulated Group, especially if the owners of the missing teeth triangulate with other people in the TG. You may as well include the iffy members anyway as you look for the common ancestor and just keep in mind that there is a chance that they don't really share the same ancestor as other members of the group. For GEDMATCH kits, sometimes a row for a shorter segment will be nearly blank because that match is not one of the top 400 matches. You can manually confirm the more distant match's ICW status by running a one-to-many in GEDMATCH for that person's kit number and comparing the names in the TG you found to the names on his one-to-many match list or by running a one-to-one with someone else in the group who does ICW the others to see if the same segment exists between them.
Other times the segments you have will "march across the page" from left to right, as in the middle example above. Consequently, the earlier (top) ones won't completely overlap the later (bottom) ones. In this example, you can see the top batch of segments form an ICW rectangle and the bottom ones do too, but some of the ones in the middle (such as the red 21.11 cM segment) are "gluing" the two ICW rectangles together. In cases like this I am inclined to believe that I have a much longer segment of ancestral DNA than any of my matches do and they are each matching me on different parts of my longer underlying (invisible) ancestral segment. I generally treat these groups as TG, although I won't be surprised to find a slightly different common ancestor in the same branch of my tree for the ones in the top formation than for the ones in the bottom. The last example above (right) is a TG embedded in another TG. When this happens you will frequently see a pattern that looks like the five spots on the face of a dice cube, as in this case. This usually means that the two segments in the center are matching one parental chromosome of the pair and the ones surrounding them are matching the other.
Here is another circumstance to be aware of. Suppose you match Fred Flintstone and Barney Rubble and they are ICW to each other. But when you go look for Fred, the common segment he shares with you is on chromosome 11 but you don't see Barney on that chromosome at all. And when you look for Barney his segment is on chromosome 21, but Fred isn't there at all. You know Fred matches Barney because the ICW tells you so. How come you don't see a segment that they both match? It's important to remember that ICWs tell you that two of your matches happen to match each other - but they do not tell you how or where they match. Often two ICWs will match each other on the same segment they share with you. But sometimes they match on another segment (or chromosome) entirely - one that you, yourself don't have and that you therefore cannot see. In most cases, when this happens, there is no common ancestor for all three of you because your two matches just happen to be related to each other in some other way. Of course, you and Fred probably still have a common ancestor, and you and Barney do too - just not the same one. This is much more likely than you might think. Notice all the isolated ICW bricks that speckle your ICW matrix? Those are instances where two of your matches just happen to be related to each other - probably in a way that doesn't involve you. In fact, this is much more likely than the alternate explanation for this situation - that two of your matches got two separate pieces of DNA from the same common ancestor - one you can see and one you can't. Multiple IBD segments from the same ancestor are relatively unlikely beyond about 3rd cousin.
Often, if you have tested several of your siblings and other close relatives and you have a flurry of short IBS segments on a chromosome you can end up with a jumbled looking ICW matrix like the one on the left below. Those long vertical stripes and rows with lots of bricks belong to siblings of the person who was tested in this example. And there is one TG embedded in another with a lot of probable IBS segments in the lower right corner that's nearly impossible to make out.

You can temporarily "clean up the mess" - just so you can see the triangulated groups more clearly - by using the Segment report and unchecking the Close relatives checkbox [FTDNA kits only] and increasing the minimum segment length to 10 cM. This is what was done for the output for the same chromosome on the right. Now you can clearly see that one 3x3 TG is embedded in another 3x3 TG in the bottom six rows of the matrix on the right. Also, be aware that you can often use your browser's back and forward buttons to toggle back and forth between the two versions of the report without having to rerun ADSA since your browser may store copies of both web pages in its cache.
Ashkenazi ICWs can be particularly difficult to visualize because there is so much interrelatedness in that community. But by tweaking the relationship filters and minimum segment size, you may be able to clear out enough of the "snowstorm" of ICWs to see some of the hidden TGs as in the example below:
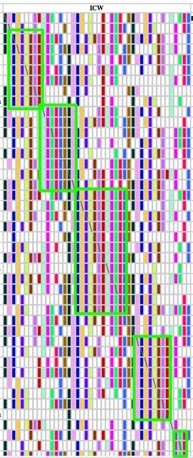
This is more of a problem with FTDNA kits than for GEDMATCH kits because the latter are limited to providing ICWs only for closer relationships (top 400 matches.)
Linking FTDNA Triangulated Groups to GEDMATCH Triangulated Groups
If you have tested at one or more testing company and have loaded your raw data to GEDMATCH you can try to combine your results using GEDMATCH and ADSA. Suppose, for example, that you have tested at AncestryDNA and FTDNA and have loaded your raw data to GEDMATCH. And suppose you found a match on Ancestry that you want to explore further. Perhaps the match has a tree that includes an ancestor you are interested in researching. If you can convince the match on AncestryDNA to transfer her raw data to GEDMATCH, then you can run a GEDMATCH One-To-One report comparing you and the AncestryDNA match to see which DNA segments you share with her (the Chromosome number and starting and ending base pair locations). You can then run ADSA on your GEDMATCH kit, supplying the chromosome number and start and end location for each matching segment to find a Triangulated Group(s) that contains that segment. If there is someone in that GEDMATCH triangulated group that has a kit number with "F" as the first character, then you can consult your FTDNA ADSA report, entering the chromosome number and the start and end locations of the triangulated group you found in GEDMATCH. If there is a triangulated group in the same location on the same chromosome in FTDNA, and it contains one or more of the people that you found in the GEDMATCH triangulated group, then you might reasonably presume that the two groups are actually one, large group and share a common ancestor. This presumption is made stronger if you have more than one "linking" person in the two groups and you have confirmed that they triangulate with you in both groups.
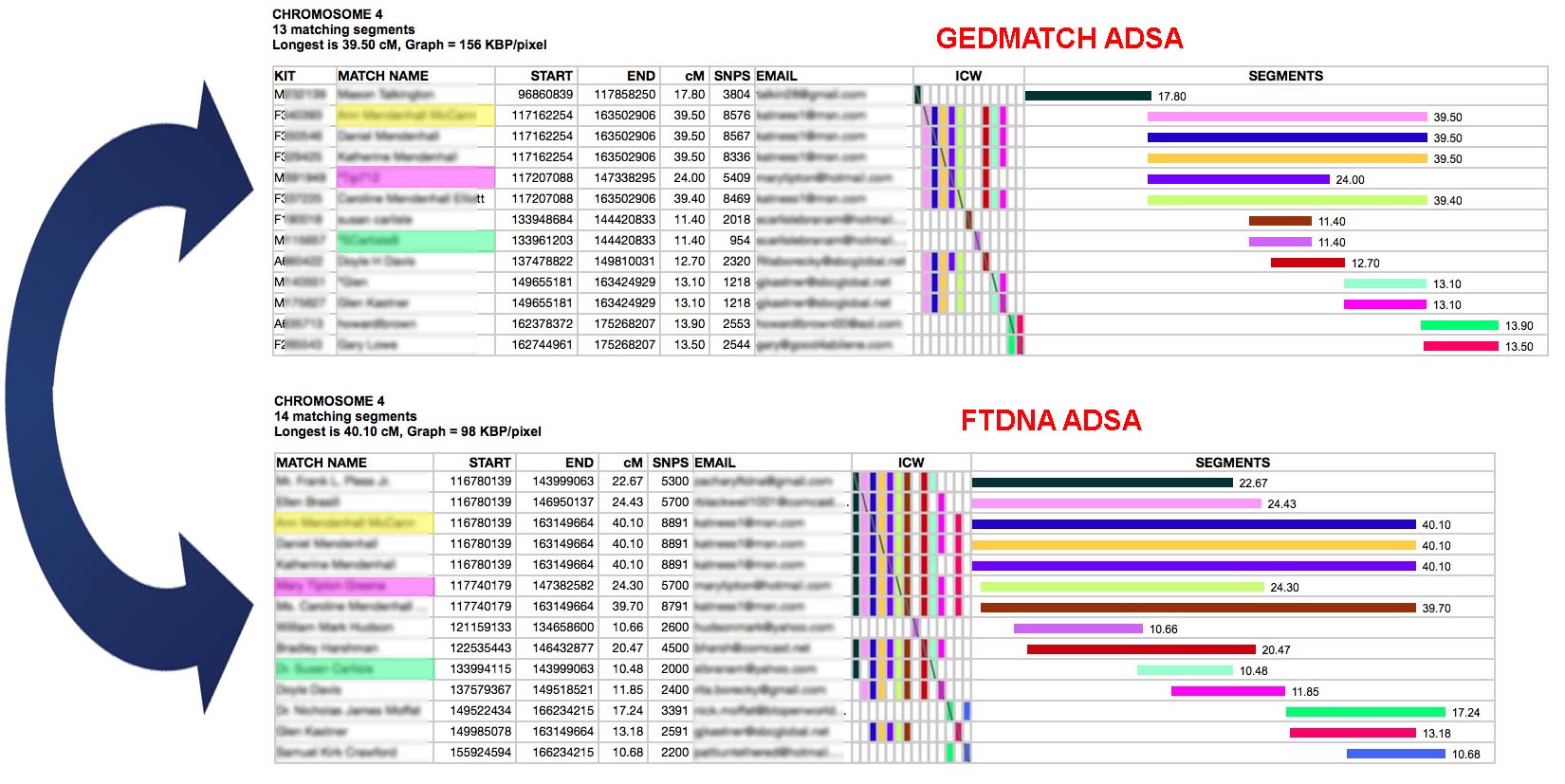
Consider the above example. The top table is an ADSA report on a GEDMATCH kit for Chromosome 4, Base Pairs from 117,500,000 to 175,000,000. So, only a portion of the chromosome is shown. The bottom table is an FTDNA kit for the same chromosome and locations. By comparing them I discovered some people who appeared in both - highlighted in yellow, pink and green. The segment lengths are slightly different because GEDMATCH and FTDNA use different algorithms for comparing segments. In the green match's case, in GEDMATCH their segment was not long enough to be considered part of the top 400 matches - so she did not get any ICWs and appears not to triangulate with the rest of the group. However, she does triangulate in FTDNA because they provide ICWs for all matches. Also, I was able to confirm that she has a matching segment to others in the GEDMATCH group by running the one-to-one tool between her and the pink and yellow matches in GEDMATCH. It would seem that I can consider everyone in the top triangulated group and everyone in the bottom one as one, larger triangulated group together. Matches who do not ICW others in their group (nearly blank ICW rows in the formations of colored bricks, or do not share the segment with others in the one-to-one tool) are either IBS matches or they match on the other parent's chromosome of the chromosome pair.
Next Steps
Once you have identified and selected a confirmed TG, it's time to try to find the common ancestor. With FTDNA kits you can use the Ancestral Surnames listed in the pop-up window for each of the matches in the TG to look for commonalities. Look for surnames you know you have in your tree, or even just surnames you don't recognize but that appear in more than one of your TG matches' trees. Failing that, look for common locations and time periods, if provided. If you have members of the group that are known close relatives, you will be able to narrow the hunt to a portion of your tree.
One feature I have used successfully in identifying common ancestors in triangulated groups with FTDNA kits is to use the surname highlighting in the SEGMENTS report type in ADSA:
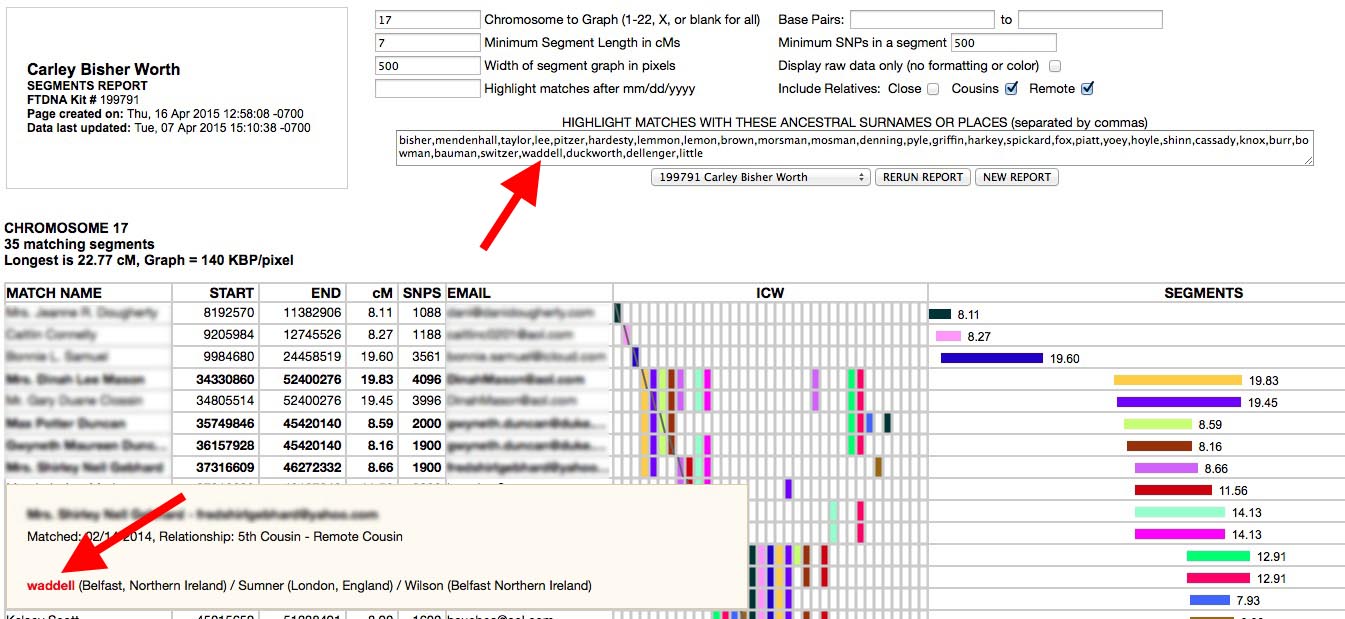
You can create a list of all your ancestral surnames (or even just the ones on one side of your family or the other) separated by commas in any editor and copy and paste the list into the SEGMENTS report surnames box. ADSA will then highlight rows for matches who have any of the names you listed in their Ancestral Surnames. The rows with matching surnames will be in bold face and the surnames found will appear in red in the pop-up windows. This works for locations too (eg. "North Carolina,NC,Texas,TX"). Note that this feature only works with Family Tree DNA kits. GEDMATCH kits do not have a surnames field.
Finally, once you've done the best you can to come up with a commonality on your own, it's time to contact the members of your TG by email. You can use ADSA to get their email addresses, then assemble them into a single group email. Ask them to REPLY ALL to your message so that everyone in the group can be in on the conversation and work together. Give them an overview of the major locations and time periods in your tree (eg. "My mother's family came to California from Canada in the 1910s and from Pittsburgh to California in the 1880s.") Sometimes these can be more useful than actual surnames. Attach a list of the surnames of your direct ancestors going back at least 7 or 8 generations. If you have your genealogy on a public web site, include a link to it. You may also want to create an ADSA report for the chromosome where the TG occurs and SAVE AS the page to a file and include that file as an attachment to your email. Be sure to keep separate folders on your computer for each TG and store copies of your correspondence and any file attachments exchanged by group members.
If you manage to find the common ancestor be sure to record what you found so that later matches that are added to the group can be quickly identified. Hopefully you will learn something new to add to your paper genealogy in the process (that's the whole point of this after all!) You may also want to use a graphical genome mapping program to create a chart of your chromosomes, labeling each ancestor to his segment of DNA.
Stack the Deck
One way to greatly facilitate finding common ancestors is to test your close relatives. Testing one or both of your parents is the best (if they are living) because it allows you to use third-party tools to pseudo-phase your data and quickly separate matches to your father's side of the family from your mother's and vice versa. If your parents are not living, test aunts or uncles - you could share 25% of their DNA with them and any of your matches who match your paternal aunt, for example, will be on your father's side of the family. Testing siblings is a good way to add more matches to evaluate. Because of the way DNA recombination works your brother or sister will very probably have some matches that you don't and vice versa. And you should test known 1st or 2nd cousins so that you have a basis for comparison to help you narrow down the portions of your tree that you need to search. Remember, though, that while a match to a close relative (other than your parents) may indicate where to look for a common ancestor, the lack of a match does not prove the opposite. This is because you only share a portion of your DNA with most of your relatives. Also, there isn't much point in testing 5th cousins, for example, for this purpose. There is only a one in ten chance that there will be enough DNA shared between you and a 5th cousin to call a match. Stick to closer relatives if you can. Most importantly, test all your oldest relatives first! There will come a time when it will be too late to do so, and older generations will give you more "reach" back into earlier times.
Be sure to post your GEDCOM file and surnames with your testing company. And, if you can afford it, test with more than one company so that you have access to each of their databases of potential matches. Don't think of it as paying for something you already bought (the test). Think of it as submitting your resume to more than one job-hunting database. Along those lines, be sure to upload your raw data to GEDMATCH.COM. It's free and it's another database of matches. And, as described earlier, you can use it to compare results between kits from different testing companies.
The DNAadoption.com Methodology
Another way to systematically process your results is to use the DNAadoption.com Methodology for Identifying your Relatives through your atDNA Results. To do this you will need to use a spreadsheet application in conjuction with the JWorks or KWorks programs here on DNAgedcom (look under the Autosomal Tools tab). These tools arrange your match data so that overlapping segments are presented as groups or sets. You will need to use your Chromosome Browser and ICW files to use this alternative method of finding Triangulated Groups (TGs).
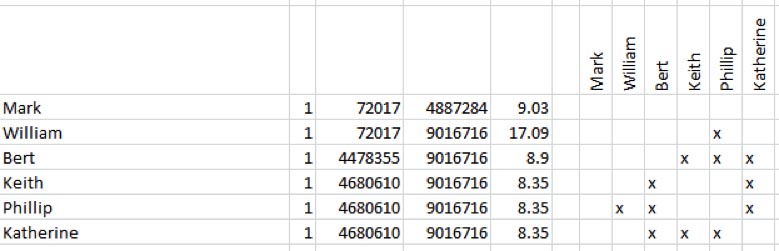
The output from JWorks and KWorks displays "X"s in the boxes of matches who are ICW in an .xls or .csv file. More information on JWorks/Kworks can be found here.
SAVING YOUR ADSA RESULTS
At any time you can save your ADSA report by using the SAVE AS... option (usually under the FILE menu) in your web browser to save a .htm or .html file to your computer's hard drive. ADSA will automatically suggest a file name with the name of the person tested and the date so that you can keep these saved reports straight. If you double-click on one of these files, you will be able to see your report again and still use the cursor hovering capabilities to see the pop-up windows. You will not be able to change the input parameters and re-run your report from these saved pages, however.
Browsers differ on how best to save these files. Unfortunately I can't give you exactly the correct option to use for each combination of browser and operating system - you will have to experiment a little. When you get into the SAVE AS... panel you will usually see a drop-down menu below the file name with the file type. Options may include "HTML file only" or "Web Archive" or "Web Page Complete" or other variations. Try SAVE AS... and use the default first. You can test it if you close your browser, then double-click on the file you just saved and see if it brings up something other than the ADSA report you were just looking at. If it didn't work right, try again and, on the SAVE AS... screen in the little drop-down menu below the file name, select one of the other options. I have found that an option may work correctly with one browser but does not work correctly with other browsers. As far as I know, "Web Archive" never works. If you're using Mac Safari, make sure it says "Page Source" and not "Web Archive".
Expert Mode
This section really gets down in the weeds! If you are a newbie, you can safely skip it and just use the canned reports described earlier. But if you have all that down pat and want to do more with ADSA, here we go... (Note: A few examples of Report Descriptions you might be interested in are provided below.)
Other than the kit number of the test you want to use as input, Expert Mode has only one other input field: the Report Description. But you can do almost anything you can think of with that one field. Examples of different Report Descriptions are included at the end of these instructions. Also, once you have set up your own custom Report Description, there is the option to SAVE it so you can use it again.
When you first see the Expert Mode input panel this is what is in the Report Description:

This looks pretty scary at first, but I will walk you through it. This default report description does exactly what the first version of ADSA was "hard wired" to do. The difference now is that you can customize it to do precisely what you want. If you haven't already, before you delve into these instructions run ADSA on your data with the Classic ADSA report first just to see what that report looks like and how it works.
Now that you've tried ADSA out, here is what the default Report Description means:

The first word describes the type of report. It can be either Segments, meaning every row describes a segment, or Matches, meaning every row describes a person who matches you. With some people in your match list, you may match them over multiple segments; however, for any one segment, you only have a single match (person) associated with it. Therefore, Matches Reports can only display fields associated with people, whereas Segment Reports can display either fields that are associated with segments or with matches.

The layout of a typical row in your report immediately follows the report type word. This is where you list the fields you want to appear in the row. In this case the report columns from left to right will be name, start, end, cm, snps, and email, followed by the "In Common With" (ICW) matrix and the segment graph (the graph will be 500 pixels wide) .

If you put a list of fields and quoted strings in parentheses following a field name, that defines a pop-up window that can be viewed by hovering the cursor over that field. In this case, if you hover over the name field (here the field is "name", which is your match name) you will see this:
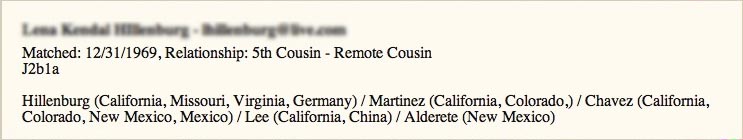
Following along the Report Description above, the pop-up contains: the Name field, a hyphen (with spaces either side created by what is in quote marks), and the Email field [these are blurred out here], then there is a new line (created by the forward slash), then Matched: (created by the quote marks), then Date (which is a field), then ", Relationship: " (which has been created by what is in quotes), then Range (which is a field for the relationship range), then a new line (created by the forward slash), then Haplogroups (which is a field for Y-DNA and/or mtDNA haplogroups), then 2 new lines created by 2 forward slashes, then Surnames (which is the list of ancestral surnames) - all of this is in a pop-up, as it has been enclosed in parentheses:

After the layout of fields in the report, you must tell ADSA how to sort the rows. In this case, it will first sort them by the chromosome number of each segment. It will also separate the rows for each chromosome into their own table and will construct a separate ICW matrix for each chromosome as well. It will then sort the rows for a given chromosome by the start location of each segment, then by the end of each segment, and finally by the match name. If you do not include table in the sort phrase, then you will get one big table with all the rows in it. If you do not include matrix you will get one big ICW matrix. You can also add words after a field name to sort ascending or descending (low to high or high to low) - if you don't specify, ascending is the default. For example, the following Report Description displays all your matches in one big table and an ICW matrix with the people with the most shared cM first: matches name shared matrix sort shared descending.

Finally, the Report Description can have a where phrase to tell ADSA which rows to select for display. There are usually hundreds of segment rows for each chromosome, so it's important to tell ADSA which ones can be skipped or else your tables will be so big that your browser will be unresponsive (and you will be looking at an awful lot of IBS segments). In this case, only rows for segments that are at least 7 cM in length and contain at least 500 SNPs will be displayed in the report.
Now try editing the Report Description and running ADSA again. For example, change it to limit the report to just one chromosome by adding this to the end of the where phrase: and chromosome=2 . Or tell it also to include the chromosome number in the rows by inserting the word chromosome before start in the layout portion. You can also add a highlight phrase after the where phrase. It works just like the where phrase, but the selected rows are bold-faced. (Highlighted rows are selected from the rows chosen using the where phrase.) For example, try adding another line that says highlight shared > 50 . You should see rows in bold for people who share more than 50 cM across all your chromosomes with you.
I have tried to make the "language" ADSA uses for Report Descriptions as flexible as possible. You can use punctuation or go to a new line in the Report Description to make things look clearer to you, but ADSA will ignore all that. It also doesn't care about capitalization and you can use either double or single quotes (as long as you use the same type to define the start and end of a string). You can also abbreviate words as long as your abbreviations are unique. For example, believe it or not, "se na so ch w cm > 7" is all you need to produce a very simple, but not particularly useful report! And there are several "noise" words that ADSA will allow you to include to make the definition seem more like English, but that it will ignore. For example, "is at least" is the same as "least" - the "is" and the "at" are not really needed and are ignored, or you could say "sort by chromosome" and the word "by" will be ignored.
Note there is an option to SAVE your own custom Report Descriptions to reuse again. You can make up a name in the blank next to the SAVE button and ADSA will save whatever is in your Report Description box on the server by the name you created. The drop-down menu next to the LOAD button will update to show anything you save. If you want to replace a saved Report Description, you can load it, then save it again with the same name. If you save a blank Report Description, the saved one you name will be deleted.
Here is ADSA's complete vocabulary:
TYPES OF REPORTS
| segments | List a segment on each row |
| matches | List a match (person) on each row |
PHRASES
| The Layout Phrase immediately follows Type of Report. (You need not start with "layout", but you can if you want to.) | |
| sort | Start the Sort Phrase (and end the layout phrase) - describe how to sort the rows (you must have a sort phrase) |
| where | Start the Where Phrase - describe how to select which rows to display (the where phrase is optional) |
| highlight | Start the Highlight Phrase - describe how to select which rows to bold-face (the highlight phrase is optional) |
FIELDS (can be used in LAYOUT, SORT, WHERE or HIGHLIGHT phrases)
| kit | Kit number (GEDMATCH) or unique ID number (FTDNA) |
| name | Name of match |
| date | Date of the match |
| range | Range of predicted relationship (eg. "3rd Cousin - 5th Cousin" ) |
| relationship | Predicted relationship (eg. "2nd Cousin") |
| shared | Total cM shared with match |
| block | Longest block shared with match |
| known | Actual relationship (if known) (eg. "Mother") |
| Match's email address | |
| surnames | Ancestral surnames of match (eg. " Smith / Jones / Brown / Taylor " ) |
| haplogroup | Haplogroups (Y-DNA and/or mtDNA) of match |
| notes | Notes you made about this match |
| allsegs | List of shared segments by chromosome and size for this match (eg. " 3 (1.5) 7 (8.3) " ) [layout only] These only include segments that have been selected for this report. |
| icws | List of matches who are in-common-with matches with this match (eg. " George Jones / Beth Smith " ) [layout only] These only include ICWs with other matches selected for this report. |
| chromosome | (Segments report only) Chromosome of this segment |
| start | (Segments report only) Start of segment in Base Pairs |
| end | (Segments report only) End of segment in Base Pairs |
| cm | (Segments report only) Length of segment in cM |
| snps | (Segments report only) Number of SNPs in the segment |
SPECIAL "WORDS"
| matrix | (Layout only) Display a row of the "In Common With" (ICW) matrix at this point in the layout |
| graph 500 | (Layout only) Display a row of the segment graph (optional width in pixels) at this point in the layout |
| / | (pop-up window definition only) Go to a new line |
| raw | (Anywhere) Display table(s) without colors or graphics so they may by copied and pasted to Excel (segment graph will be skipped). |
SORT words (follow field names in Sort Phrase)
| ascending | Sort the preceding field from low to high (default) |
| descending | Sort the preceding field from high to low |
| table | Create a new table each time the preceding field's value changes from one row to the next |
| matrix | Start a new ICW table each time the preceding field's value changes from one row to the next |
COMPARISON OPERATORS (for WHERE and HIGHLIGHT phrases: field operator value)
| contains | preceding field contains the following string - eg. surnames contain 'smith'. You may use % and _ as wild cards if you wish. % matches any number of characters, _ matches one. So "fred%kson" matches "Frederickson" and "Fredrickson". Also, you may have more than one string to match, eg. 'smith,jones,webber' in CSV (comma separated values) format. Multiple strings imply that the field will match any of them (smith OR jones OR webber). Upper and lower case is ignored. Spaces are matched (eg. "mendenhall, taylor" will match "mendenhall" and " taylor") Do not use CONTAINS with numeric fields or dates. |
| equals, =, == | preceding field equals the following value (number or string) - eg. chromosome=17 |
| more, greater, beyond, after, above, longer, > | preceding field is greater than the following value (number or string) - eg. shared above 50 |
| least, >= | preceding field is greater than or equal to the following value (number or string) - eg. cm at least 7 |
| less, before, below, < | preceding field is less than the following value (number or string) - eg. block less than 50 |
| <= | preceding field is less than or equal to the following value (number or string) - eg. start <= 40000000 |
| <> | preceding field is not equal to the following value (number or string) - eg. name <> "George Jones" |
LOGICAL OPERATORS (for WHERE and HIGHLIGHT phrases)
| and, & | Logical "and" - eg. (cm > 7) and (snps > 500) |
| or, | | Logical "or" - eg. surnames contain 'smith' or surnames contain 'jones' |
| not | Logical "not" - eg. not surnames contains 'martin' |
| ( ) | Parenthesis may be used in the WHERE and HIGHLIGHT phrases to clarify comparisons - eg. ((cm>7 & snps>500) or (not shared < 1000)) |
EXAMPLE COMPARISONS
| cm>7 and snps>500 | Pick rows where segment is greater than 7 cM and has more than 500 SNPs |
| shared <= 400 | Do not display rows for close relatives (those with more than 400 cM shared DNA) |
| date after '1/25/2014' | Pick only rows for matches that were added after 1/25/2014 (American date format) |
| surnames contain 'martin' | Pick rows for matches who have the surname "Martin" among their Ancestral Surnames |
| not known contains "mother" | Do not pick rows that have "mother" in the known relationship field |
| surnames contain 'england,france' | Pick rows for matches who have "England" or "France" as places among their Ancestral Surnames |
| start < 140000000 and end > 119000000 and chr=2 | Pick only segments that overlap the base pair range from 119000000 to 140000000 on chromosome 2. |
THE FOLLOWING WORDS AND CHARACTERS ARE IGNORED (UNLESS IN A QUOTED STRING VALUE)
<extra blanks, new lines, etc.> , . ; : * % then than cousin by to at is layout
Here are a few example Report Descriptions:
| List all matches from most shared cM to least. Highlight rows with "Jones" as a surname. | matches |
| Create ICW matrix of closest matches | matches |
| Standard segment report, highlighting recent matches | segments |
| Highlight rows for matches who have the Ancestral Surnames "Taylor" or "Smith" | segments name (name ' - ' email / 'Matched: ' date ', Relationship: ' range / haplogroups / / surnames) start end cm snps email matrix graph 500 (name ' - ' email / 'Matched: ' date ', Relationship: ' range / 'Total Shared: ' shared ' cM, Longest Block: ' block ' cM' / 'Segments by Chromosome (cM): ' allsegs / / icws) Sort chromosome table matrix, start, end, name Where cm>=7 and snps>=500 and Highlight surnames contain "taylor,smith" |
| Exclude all close relatives through 1st cousin to reduce clutter in the ICW matrix EXCEPT my 1st cousin, Fred Flintstone. Highlight his segment rows in bold to make comparisons in TGs easier. | segments |
| This is the Report Description used by the Classic ADSA and Segments reports for GEDMATCH kits | Segments |
Read More About ADSA
- "Introducing the Autosomal DNA Segment Analyzer", DNA-eXplained, Roberta Estes, 9 Jan 2014
- "Autosomal DNA Segment Analyzer (ADSA) - No Spreadsheets Required!", Genealogy Junkie, Sue Griffith, 15 Jan & 21 Feb 2014
- "The Worthwhile Autosomal DNA Segment Analyzer", Family History Research by Jody, Jody Lutter, 15 Jan 2014
- "Oh, Charlie!", The Legal Genealogist, Judy G. Russell, 2 Feb 2014
- "Why Autosomal DNA Test Results Are Significantly Different for Ashkenazi Jews", AVOTAYNU, Jeffrey Mark Paull, Volume XXX, Number 1, Spring 2014
- "ADSA is Now Available for GEDmatch Kits!", Genealogy Junkie, Sue Griffith, 13 April 2015
READ THIS IF THINGS DON'T WORK FOR YOU TROUBLESHOOTING TIPS The following are a few things that have been reported to support@DNAgedcom.com. Your problem may be among them. When downloading data with DNAgedcom.com... There are troubleshooting tips in the data download instructions here:
After you click the GET REPORT button on the ADSA input screen... If you click on GET REPORT and, after a while, you get a blank screen or a 500 Server Internal Error or something of the kind, it means the server timed out trying to retrieve your data from the database or constructing the report. In some cases (such as if you have Ashkenazi ancestry) you may have far too many segments and ICWs for ADSA to handle gracefully. Unchecking the Remote Relative input checkbox for the Segments report is also a way to make ADSA run faster with less possibility of a timeout. Or you can limit the report to just one chromosome at a time. For very large datasets, avoiding highlighting surnames or dates can also speed things up. Unbalanced ICWs Sometimes you will get a warning message that a number of your ICWs are unbalanced. An unbalanced ICW is where you got "Fred matches Mary" (where Fred and Mary are two of your matches - Fred had Mary as a match in common with you) but you didn't get "Mary matches Fred". Ideally, you should always get both sides of the pair. Having unbalanced ICWs can mean your download may have ended before getting all the ICWs downloaded, or FamilyTreeDNA might not have sent all the ICWs because of a glitch on their end, or the downloader ran out of memory while gathering your ICWs. Your unbalanced ICWs may not be significant as long as your last download completed successfully and there were no errors in an error log. The way to tell is to look in the table that is on the Family Tree DNA downloader page in DNAgedcom. Make sure the Date Last Completed date/time for this kit is AFTER the Date Last Loaded date/time. If it is, the downloader thought it finished. Also, look in the VIEW FILES screen under the MEMBERS menu item for an error log file for that kit and download it and open it in Excel or a text editor. If there are no errors, that's a good sign. Finally, check the "not matched" counts in the table on the downloader page - the ones for Chromo (Segments) and ICW should be low or zero. A few in Matches not matched is OK - it could just be people without any ICWs (which is possible in some cases). Often, even if you get an unbalanced ICW warning, you will still see all your ICWs in the ADSA report because ADSA can infer the missing ICW bricks from the ones already present in the data you downloaded. These appear with white Xs on them. If none of this solves your problem, feel free to email me at worth@ucla.edu and we'll figure it out together
Here are the error messages that are possible and an explanation of their meaning:
|
ASP.NET Web Hosting By Arvixe
